At 2 a.m. one night last April, Michael Schoof triple-checked the numbers on his screen, took a deep breath, and fired off an email he’d been waiting all day to send.
“I think it’s working” was the cautious wording of his message.
Schoof, a graduate student in the lab of Peter Walter, PhD, a renowned scientist specializing in protein sorting and cellular membranes, was part of a small team on a quixotic mission: to immobilize SARS-CoV-2, the novel coronavirus that causes COVID, by using a synthetic version of tiny antibodies originally discovered in llamas and camels. These “nanobodies,” as they’re known, had come from the UC San Francisco lab of Aashish Manglik, MD, PhD, an up-and-coming protein scientist who had spent the previous three years building a vast library of nanobodies and developing new ways to exploit their unusual properties.
During the previous month, Schoof had spent most of his waking hours cloistered in the otherwise empty lab complex on UCSF’s Mission Bay campus. It was the height of COVID’s spring 2020 surge, and only essential health care staff and those working on science related to the pandemic were allowed into the University’s facilities. Schoof had dragooned his roommate, a fellow grad student named Reuben Saunders, into working with him on the project. Subsisting on steamed dumplings and gallons of tea, they had been sorting through the 2 billion nanobodies in Manglik’s library in the hope of identifying a molecule capable of glomming on to the deadly SARS-CoV-2 and immobilizing it. Now, finally, Schoof was convinced they had achieved their first big breakthrough.
The first step in any viral infection is a cellular hijacking. To gain control of a human cell, SARS-CoV-2 latches the grappling-hook-like spikes on its own exterior to proteins called ACE2 receptors on the exterior of a target cell. But what if, the researchers wondered, they could block the hijacker by giving the grappling hooks something else to latch onto?
That day, Schoof had begun running tests on hundreds of colonies of yeast, each engineered to produce certain nanobodies from Manglik’s library. All of these particular nanobodies had demonstrated an ability to latch onto SARS-CoV-2’s spikes. Now it was time to ask the key questions: How tightly had these nanobodies bound to the spikes? Were they able to compete with the ACE2 receptors?

UCSF graduate student Michael Schoof’s work was integral to the team’s success. Photo: Steve Babuljak
To find out, Schoof had mixed his nanobody-expressing yeast cells with fluorescent SARS-CoV-2 spikes. When he looked at the results from the first two plates, he felt a rush of excitement, which he quickly tempered with the scientific skepticism he’d been taught to cultivate. Some of the nanobodies were sticking to the SARS-CoV-2 spikes but could still be elbowed aside by an excess of human ACE2 receptors: clear evidence of a potential neutralizer.
“That,” he recalls, “is when we knew we had something.”
In the days that followed Schoof’s circumspect late-night email, Walter and Manglik tapped their respective networks of scientific contacts, calling in reinforcements from labs across campus and as far away as Paris to aid in the next stage of their search. Soon, the tiny team had morphed into a veritable army of cross-disciplinary researchers and graduate students. In November, they published their results in the prestigious journal Science. In the paper, almost 60 co-authors described a bold, innovative COVID countermeasure, proposing that their nanobodies could be used in an inexpensive, easy-to-transport nasal spray capable of neutralizing SARS-CoV-2. Among themselves, they dubbed the molecules AeroNabs.
This is something that you could take after testing positive that could diminish your viral load immediately.”
Since then, the UCSF team has been seeking an industry partner willing to bankroll the costly and rigorous clinical trial process, but currently pharmaceutical companies are focused on vaccine development for prevention and more traditional antibodies for treatment.
But the nanobody approach is promising. Due to the simple structure of nanobodies, AeroNabs could be far cheaper and faster to mass-produce, far easier to transport, and far easier to store than the traditional antibodies currently in use and under development.
“This is something that you could take after testing positive that could diminish your viral load immediately,” Walter says. “So your chances of developing severe disease would be reduced by this treatment.”
He also notes that mass vaccination will take time, and that not everyone in the population can or will be vaccinated, making passive protection still incredibly valuable. “And,” he adds, “we don’t know how widely the vaccine will be available beyond the world’s richest countries.”
Dynamic duo
The seeds of the AeroNabs project were planted in 2017, when Walter heard Manglik deliver a talk on his work.
At first glance, the two scientists appear to be an odd couple. With his full head of dark hair, boyish smile, and clean-shaven chin, Manglik could be mistaken for a graduate student. In fact, he’s a rising star in his field who in 2013 made Scientific American’s “30 under 30” list. Born in India, Manglik spent his first eight years in Saudi Arabia before his family immigrated to Des Moines, Iowa, where he discovered science in college. The 60-something Walter, on the other hand, sports a full white beard and mustache and small-lensed spectacles. He was born and grew up in Germany, came to the U.S. for his graduate work, and has charted a legendary career. His many honors include the prestigious Lasker Award, often seen as a precursor to a Nobel Prize. But despite their differences, Walter and Manglik share a profound passion for molecular biology and its endlessly flexible organic building blocks: proteins.

Peter Walter (left) and Aashish Manglik (right), photographed at UCSF’s Mission Bay campus, share a fascination with proteins. Photo: Noah Berger
Manglik’s talk that day was about his effort to assemble one of the world’s largest libraries of nanobodies – a promising, relatively new type of antibody derived from the blood of llamas, camels, and other animals in the camelid family. He had first learned about nanobodies in graduate school at Stanford, after falling in love with the study of receptors, a broad family of proteins involved in intercellular signaling. Receptors stick out of cells like antennae, each one responding to a specific chemical signal. While studying human adrenaline receptors, Manglik made extensive use of nanobodies, which, thanks to their tiny size, can interact with receptors with far more precision than the custom-made antibodies he was using to explore receptor properties. His experiments revealed how different geometrical configurations of receptors influence their signaling behavior.
“Proteins are not just simple Legos that fit together – they are like Legos made out of jello or putty,” Manglik explains. “They’re constantly moving. In fact, it’s the movement of a protein, it turns out, that really matters for how it works. And nanobodies can help us control that movement.”
Nanobodies: A Boon for Science
Nanobodies were discovered in the late 1980s by a pair of undergraduates at the Free University of Brussels, after they famously approached their biology professor, an immunologist named Raymond Hamers, to complain about an assignment. History has obscured the reason for their complaint; one widely cited account holds that the students were worried that the assignment, which required them to analyze the antibodies in human blood, might infect them with a disease. Another version has it that the students felt the experiment was boring and asked their professor to assign them something more original.
Whatever the truth, no one disputes what happened next. Rummaging around in a laboratory refrigerator, Hamers found a vial of frozen dromedary camel serum infected with parasites thought to cause African sleeping sickness. He gave it to the students and suggested that they isolate the antibodies in the camel blood to see what they looked like. When the students purified the blood, they discovered something amazing.
In addition to the standard antibodies found in all vertebrates, the purified samples contained a derivative antibody never before seen in science – smaller, simpler proteins, which the students at first mistook for fragments of conventional antibodies. Further examination revealed them to be an entirely new class of immune agents, lacking one of the protein chains found in all other previously studied antibodies.

Photo: Grafissimo
The discovery led to a groundbreaking 1993 paper in the prestigious journal Nature. Hamers and his students dubbed the new diminutive proteins nanobodies. Similar single-chain antibodies were later identified in llamas, alpacas, guanacos (another long-necked South American mammal), and even sharks.
It soon became apparent not only that nanobodies were useful immunologically, but that their small size made them useful experimental tools – as Manglik and his UCSF colleagues can amply confirm.
Studying how these gelatinous molecular-level building blocks move, snap and unsnap, and interact became Manglik’s focus when he joined the UCSF faculty. He knew early on that nanobodies would be a big part of his work. Though antibodies and nanobodies exist to help animals fight off infection, Manglik also sees them as an endlessly malleable tool that can be used to hack into a wide array of processes in the human body as well as decode basic scientific mysteries. But nanobodies were time-consuming to make and required access to camelids. As a graduate student, Manglik had relied on a collaborator in Belgium who would inject a receptor protein of interest into a llama, then harvest the nanobodies from the animal’s blood. The entire process took months of very specialized work, which only a few groups had the capability to do.
To democratize access to nanobodies for researchers everywhere, Manglik teamed up with Andrew Kruse, PhD, a close friend from grad school who had joined the faculty at Harvard Medical School. Together, the two labs created trillions of unique nanobody-encoding DNA sequences, each inspired by the nanobodies normally found inside llamas. The DNA sequences for these nanobodies are housed in a vast pool of billions of diminutive yeast cells, each of which can be coaxed to put a copy of an individual nanobody on its surface. Completely bypassing the need for a living llama, such a library gives researchers access to yeast cells harboring nanobodies specific for any given task. Manglik and Kruse have openly shared their libraries with hundreds of labs around the world.

UCSF graduate student Bryan Faust examines a 3D representation of a SARS-CoV-2 spike protein, created using a cryo-electron microscope.
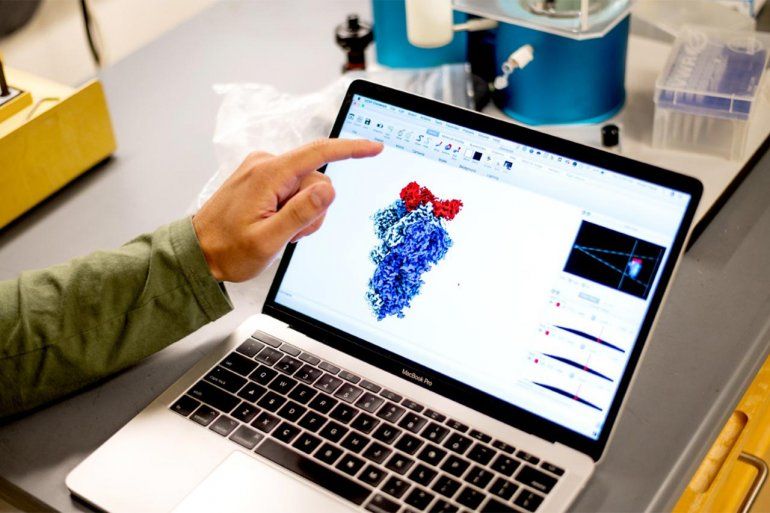
The cryo-electron microscope enabled the team to visualize the nanobody-spike protein interface. Photos: Noah Berger
“The idea is that in an animal, there are trillions of different nanobodies to fight against anything that it can encounter,” he says. “We wanted to make a library that encoded for billions of individual nanobodies. This library would be a great starting point for finding a nanobody against basically anything – all in the lab and without the need to inject an animal.”
After hearing Manglik explain all this, Walter steered his graduate student Michael Schoof to Manglik’s lab. Schoof was trying to modulate the behavior of a protein related to traumatic brain injury, and Walter suspected that Manglik’s nanobodies might be useful in that effort.
Then the coronavirus hit, the world stopped, and nearly all non-COVID-related activity at the University shut down.
“So at that point, we said, ‘Well, we can either sit at home now, or we can think how we can really help in this push for a solution,’” Walter recalls.
Within a few days, Walter and Schoof were in email contact with Manglik. They knew the disease-fighting properties of nanobodies. A nanobody technology had recently won FDA approval to treat a blood-clotting disorder, and another one, used to treat a respiratory virus, had reached late-stage clinical trials.
Was it possible they could build one to fight the coronavirus?
An amazing result
From the beginning, the team knew, the success of the project would rest on their ability to find a nanobody with sufficient binding affinity – the ability to attach to and straitjacket the coronavirus’s spikes.
Proteins have specific shapes. How well two proteins fit together determines their binding affinity. Walter and Manglik knew that the binding affinity that causes SARS-CoV-2 to adhere to ACE2 proteins could theoretically be overpowered by a nanobody shaped in just the right way.
Manglik already had a key ingredient for such an experiment. Researchers at the University of Texas (UT) at Austin had recently revealed the unique structure of the SARS-CoV-2 spikes which allowed the virus to bind to human cells’ ACE2 receptors. Manglik reached out to UT’s Jason McLellan, PhD, who agreed to send him their “construct” – a piece of DNA coding for the spikes that could be inserted into another cell, expressed in large quantities, purified, and used for experiments.
The team began screening the 2 billion nanobodies in the library to see if they could find compounds with the right binding affinity to the SARS-CoV-2 spikes. Within three weeks, they’d identified 800 potential candidates, and a week later Schoof wrote his cautious late-night email informing Manglik and Walter that he’d seen some initial positive results. By late April, the team had identified 21 distinct nanobodies that seemed to compete with the ACE2 receptor, theoretically blocking the SARS-CoV-2 attachment mechanism.
That’s when the tiny team began to supersize, recruiting structural biologists to zero in on how the nanobodies bound to the SARS-CoV-2 spike protein, and then using this information to design modifications to make them even more powerful.
That required purifying 21 candidate proteins, testing their binding, and then using UCSF’s cryo-electron microscopy facilities to image at near-atomic resolution the most promising candidates, while they were bound to the SARS-CoV-2 spike. To complete this monumental task, they joined forces with a parallel effort known as the QCRG Structural Biology Consortium – an assembly-line-like process put together by 12 UCSF faculty members and over 60 trainees to tackle SARS-CoV-2. The effort was fueled by a sense of urgency, and the participants worked grueling hours late into the night.
Once the team had images of the top nanobodies bound to the SARS-CoV-2 spike, they began to examine each nanobody’s unique binding mechanism and used that information to design a next-generation version. They settled on constructing a three-armed nanobody consisting of three copies of a single nanobody stitched together so it could bind simultaneously to the three separate arms that make up each coronavirus spike.
After stitching together the nanobodies and testing them, Bryan Faust, a graduate student in Manglik’s lab, delivered the next exciting finding: Each of the three arms enhanced the binding of its neighbors exponentially. The ability of the improved version to bind to the viral spikes increased two-hundred-thousandfold.
“This was an amazing result – to see this huge order of improvement,” Walter recalls. “It was absolute celebration time.”

The nanobody that the team engineered could be used in an inexpensive, easy-to-transport nasal spray or inhaler. Illustration: Kouzou Sakai | Folio Art
To test the compound against a live virus, the team needed a laboratory with a Biosafety Level 3 (BSL-3) designation. The group recruited Marco Vignuzzi, PhD, a former UCSF postdoc who runs a BSL-3 lab at Institut Pasteur in Paris. By June, one of Vignuzzi’s postdocs was running the UCSF nanobody against actual SARS-CoV-2 to see if it was capable of neutralizing the virus.
The end result was both highly effective and stable – so stable that it can be delivered in aerosol form using a mesh nebulizer that Manglik purchased on Amazon.
With Big Pharma laser-focused on developing vaccines and traditional antibodies, finding a quick path to commercialization has proved challenging. But Manglik, Walter, and their team are undeterred.
“It’s almost certain that there will be more respiratory pandemics in our lifetime,” says Manglik. “It could be influenza, anther SARS pandemic, or some pathogen we don’t even know about yet. For the next pandemic, the hope is that researchers could go not only as fast as we did, but maybe even faster.”
Without doubt, it would be hard to find a more potent testament to the delightful unpredictability and potential of modern science – that a pandemic which has caused loneliness, suffering, and death also gave rise to this eclectic crew and their potentially lifesaving solution that just a few years ago might have seemed absurd.
“It’s just one of those things where you say, ‘We want to go on this adventure,’” Walter says. “We committed to it, and then it just worked much better then we could have dreamt.”